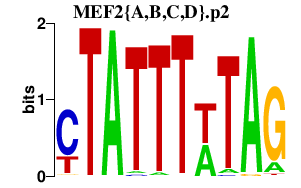
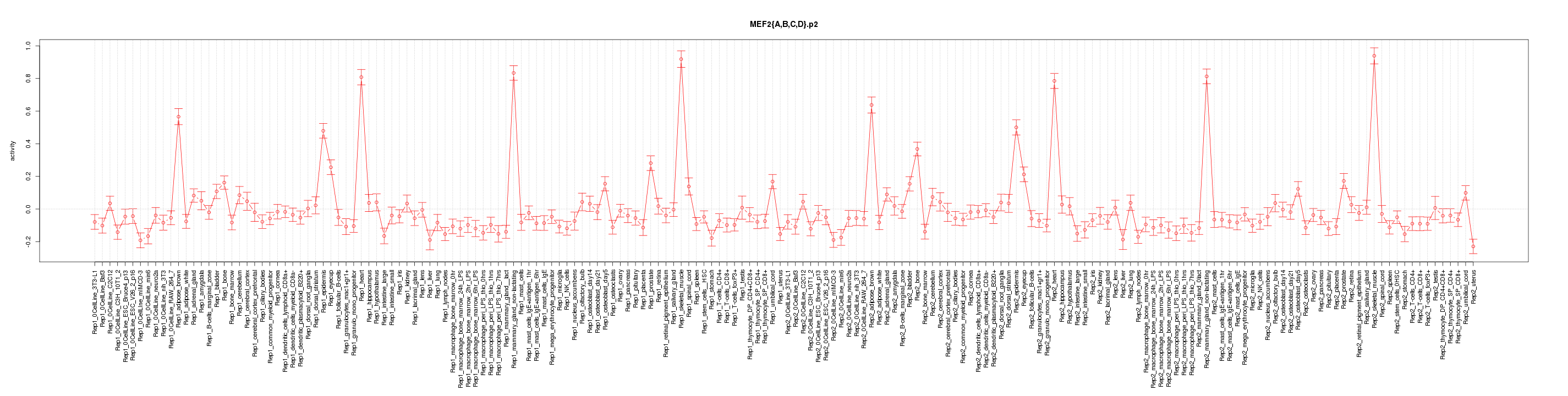
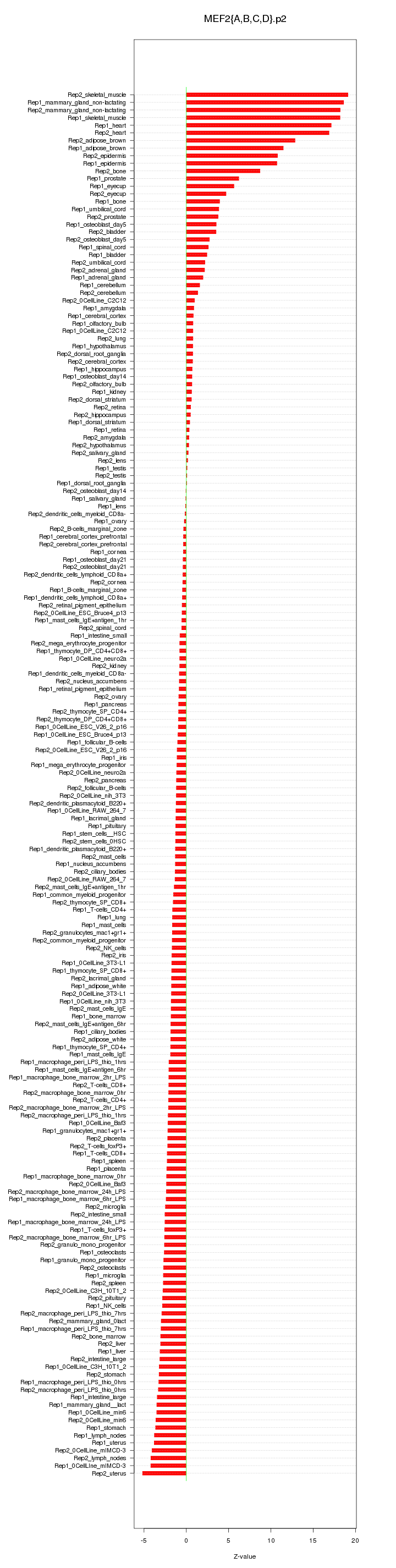
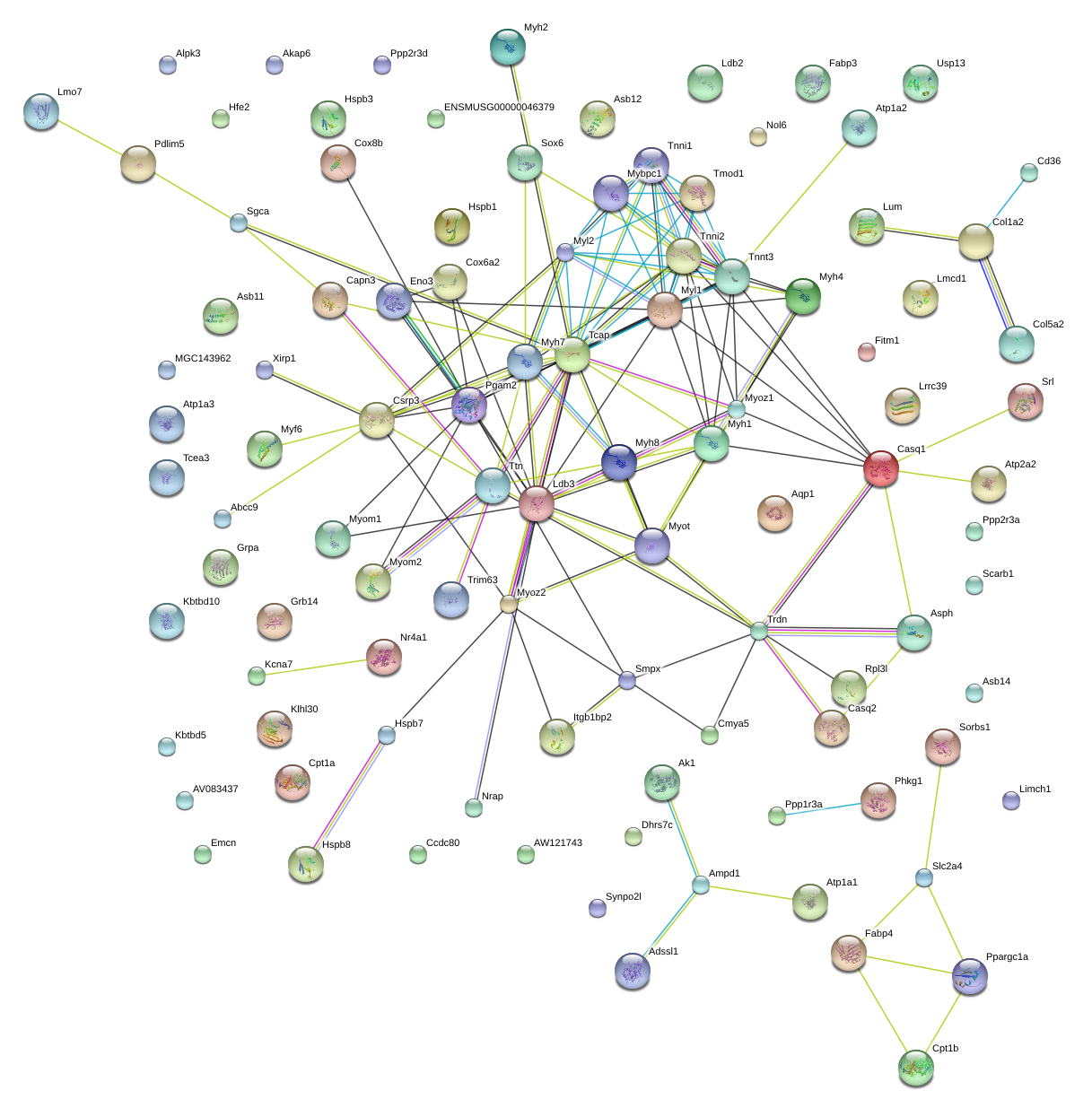
|
chr1_-_66981900
|
148.114
|
NM_001113387
|
Myl1
|
myosin, light polypeptide 1
|
|
chrX_+_154137049
|
134.514
|
NM_025357
|
Smpx
|
small muscle protein, X-linked
|
|
chrX_+_154137193
|
133.431
|
|
Smpx
|
small muscle protein, X-linked
|
|
chrX_+_154137114
|
132.268
|
|
Smpx
|
small muscle protein, X-linked
|
|
chr11_+_98245124
|
122.898
|
NM_011540
|
Tcap
|
titin-cap
|
|
chr6_-_14705104
|
119.130
|
NM_080464
|
Ppp1r3a
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A
|
|
chr1_-_66991625
|
118.044
|
NM_021285
|
Myl1
|
myosin, light polypeptide 1
|
|
chr1_-_66991580
|
115.316
|
|
Myl1
|
myosin, light polypeptide 1
|
|
chr17_+_71368860
|
79.481
|
NM_001083934
NM_010867
|
Myom1
|
myomesin 1
|
|
chr6_-_142650737
|
77.781
|
NM_001044720
NM_011511
NM_021041
NM_021042
|
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr2_+_32483909
|
76.343
|
NM_001198792
|
Ak1
|
adenylate kinase 1
|
|
chr11_+_67013553
|
72.465
|
NM_030679
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult
|
|
chr14_-_21475760
|
72.068
|
NM_021508
|
Myoz1
|
myozenin 1
|
|
chr7_+_149684731
|
71.377
|
NM_001163664
NM_001163665
NM_001163666
NM_001163667
NM_001163668
NM_001163669
NM_001163670
NM_011620
|
Tnnt3
|
troponin T3, skeletal, fast
|
|
chr16_-_4523061
|
70.126
|
|
Srl
|
sarcalumenin
|
|
chr7_-_135349828
|
67.768
|
NM_009943
|
Cox6a2
|
cytochrome c oxidase, subunit VI a, polypeptide 2
|
|
chr16_-_4523051
|
67.659
|
NM_175347
|
Srl
|
sarcalumenin
|
|
chr11_-_5703677
|
66.286
|
|
Pgam2
|
phosphoglycerate mutase 2
|
|
chr11_+_67051530
|
65.649
|
NM_010855
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle
|
|
chr17_+_24869629
|
65.482
|
NM_025425
|
Rpl3l
|
ribosomal protein L3-like
|
|
chr7_-_148086285
|
64.725
|
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb
|
|
chr14_-_21483775
|
63.478
|
|
Synpo2l
|
synaptopodin 2-like
|
|
chr7_+_149628285
|
62.029
|
NM_009405
|
Tnni2
|
troponin I, skeletal, fast 2
|
|
chr8_+_15057652
|
60.140
|
NM_008664
|
Myom2
|
myomesin 2
|
|
chr11_-_69761489
|
59.392
|
NM_009204
|
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr5_+_122550987
|
59.093
|
NM_010861
|
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow
|
|
chr7_+_149627709
|
59.009
|
|
Tnni2
|
troponin I, skeletal, fast 2
|
|
chr11_+_70471297
|
58.572
|
NM_007933
|
Eno3
|
enolase 3, beta muscle
|
|
chr2_+_32480924
|
58.451
|
NM_001198791
|
Ak1
|
adenylate kinase 1
|
|
chr3_+_96329090
|
57.659
|
NM_027126
|
Hfe2
|
hemochromatosis type 2 (juvenile) (human homolog)
|
|
chr1_-_174149963
|
57.377
|
|
Casq1
|
calsequestrin 1
|
|
chr11_+_70471347
|
56.642
|
|
Eno3
|
enolase 3, beta muscle
|
|
chr10_+_32803288
|
55.300
|
NM_029726
|
Trdn
|
triadin
|
|
chr1_-_174149989
|
55.038
|
NM_009813
|
Casq1
|
calsequestrin 1
|
|
chr19_-_56464434
|
54.796
|
NM_008733
NM_198059
|
Nrap
|
nebulin-related anchoring protein
|
|
chr2_+_69508174
|
54.002
|
NM_001081087
|
Kbtbd10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr3_-_122737855
|
52.723
|
NM_021503
|
Myoz2
|
myozenin 2
|
|
chr14_-_21475715
|
52.031
|
|
Myoz1
|
myozenin 1
|
|
chr18_+_44493726
|
51.796
|
NM_001033621
|
Myot
|
myotilin
|
|
chr15_+_97962651
|
50.893
|
|
|
|
|
chr3_+_101890431
|
50.878
|
NM_009814
|
Casq2
|
calsequestrin 2
|
|
chr15_-_89256262
|
50.389
|
NM_009948
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle
|
|
chr14_-_35401845
|
49.860
|
NM_001039071
NM_001039072
NM_001039073
NM_001039075
NM_001039076
NM_011918
|
Ldb3
|
LIM domain binding 3
|
|
chr11_+_67051128
|
49.753
|
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle
|
|
chr7_-_56103405
|
49.060
|
NM_001198841
|
Csrp3
|
cysteine and glycine-rich protein 3
|
|
chr1_-_174228105
|
48.614
|
NM_178405
|
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr15_-_89256252
|
48.225
|
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle
|
|
chrX_+_160876032
|
48.002
|
NM_026853
|
Asb11
|
ankyrin repeat and SOCS box-containing 11
|
|
chr7_-_56103389
|
47.769
|
|
Csrp3
|
cysteine and glycine-rich protein 3
|
|
chr4_+_155590035
|
47.694
|
NM_172417
|
2310042D19Rik
|
RIKEN cDNA 2310042D19 gene
|
|
chrX_+_98644459
|
46.829
|
|
Itgb1bp2
|
integrin beta 1 binding protein 2
|
|
chr3_+_102877936
|
46.702
|
NM_001033303
|
Ampd1
|
adenosine monophosphate deaminase 1
|
|
chrX_+_98644445
|
46.696
|
NM_013712
|
Itgb1bp2
|
integrin beta 1 binding protein 2
|
|
chr4_+_133871031
|
46.420
|
NM_001039048
|
Trim63
|
tripartite motif-containing 63
|
|
chr14_-_35401654
|
45.908
|
NM_001039074
|
Ldb3
|
LIM domain binding 3
|
|
chr10_-_106931783
|
45.180
|
NM_008657
|
Myf6
|
myogenic factor 6
|
|
chr10_-_88067758
|
40.373
|
NM_175418
|
Mybpc1
|
myosin binding protein C, slow-type
|
|
chr4_+_140977613
|
40.282
|
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular)
|
|
chr11_+_67611772
|
40.062
|
NM_001013013
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr5_-_116872852
|
39.804
|
NM_030704
|
Hspb8
|
heat shock protein 8
|
|
chr9_-_119932620
|
39.723
|
NM_011724
|
Xirp1
|
xin actin-binding repeat containing 1
|
|
chr15_+_101097276
|
39.059
|
NM_010444
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr5_+_136363914
|
38.971
|
|
Hspb1
|
heat shock protein 1
|
|
chr13_-_93914519
|
38.196
|
NM_023821
|
Cmya5
|
cardiomyopathy associated 5
|
|
chr5_+_136363810
|
37.974
|
|
Hspb1
|
heat shock protein 1
|
|
chr7_-_148086341
|
37.562
|
NM_007751
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb
|
|
chr5_-_51945054
|
37.551
|
NM_008904
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
|
|
chr11_-_5703752
|
37.472
|
NM_018870
|
Pgam2
|
phosphoglycerate mutase 2
|
|
chr1_+_93247592
|
37.355
|
NM_027551
|
Klhl30
|
kelch-like 30 (Drosophila)
|
|
chr5_+_136363769
|
37.323
|
NM_013560
|
Hspb1
|
heat shock protein 1
|
|
chr4_+_129985957
|
37.227
|
NM_010174
|
Fabp3
|
fatty acid binding protein 3, muscle and heart
|
|
chr6_+_55286292
|
37.089
|
NM_007472
|
Aqp1
|
aquaporin 1
|
|
chrX_+_10173557
|
36.922
|
|
|
|
|
chr4_-_73596693
|
36.867
|
NM_027104
NM_027981
|
2310002L09Rik
|
RIKEN cDNA 2310002L09 gene
|
|
chr10_+_97028134
|
36.853
|
NM_008524
|
Lum
|
lumican
|
|
chr14_+_56194658
|
36.578
|
|
Fitm1
|
fat storage-inducing transmembrane protein 1
|
|
chr14_+_56194510
|
36.407
|
NM_026808
|
Fitm1
|
fat storage-inducing transmembrane protein 1
|
|
chr14_+_56194622
|
36.317
|
|
Fitm1
|
fat storage-inducing transmembrane protein 1
|
|
chr14_-_21487489
|
36.105
|
NM_175132
|
Synpo2l
|
synaptopodin 2-like
|
|
chr7_+_88202481
|
36.029
|
NM_054085
|
Alpk3
|
alpha-kinase 3
|
|
chr4_-_9570742
|
35.843
|
|
Asph
|
aspartate-beta-hydroxylase
|
|
chr11_-_94837640
|
35.790
|
NM_009161
|
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein)
|
|
chr4_+_46052090
|
34.703
|
NM_021883
|
Tmod1
|
tropomodulin 1
|
|
chr14_+_27707789
|
34.382
|
NM_001170748
NM_080856
|
Asb14
|
ankyrin repeat and SOCS box-containing 14
|
|
chr5_-_122906470
|
34.307
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr1_-_45559866
|
33.380
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr14_-_21475679
|
33.307
|
|
Myoz1
|
myozenin 1
|
|
chr12_+_113858217
|
33.096
|
NM_007421
|
Adssl1
|
adenylosuccinate synthetase like 1
|
|
chr5_+_67137054
|
32.799
|
NM_001001980
|
Limch1
|
LIM and calponin homology domains 1
|
|
chr16_+_5885447
|
32.356
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr9_-_101101334
|
31.755
|
NM_172144
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr11_-_20729367
|
31.199
|
|
1110067D22Rik
|
RIKEN cDNA 1110067D22 gene
|
|
chr1_-_45559904
|
31.141
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr13_-_114453813
|
30.995
|
NM_019960
|
Hspb3
|
heat shock protein 3
|
|
chr5_-_17394776
|
30.570
|
NM_001159558
|
Cd36
|
CD36 antigen
|
|
chr6_+_4455686
|
30.035
|
NM_007743
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr14_+_102239673
|
29.949
|
|
Lmo7
|
LIM domain only 7
|
|
chr12_+_53800369
|
29.808
|
NM_198111
|
Akap6
|
A kinase (PRKA) anchor protein 6
|
|
chr2_-_64860728
|
29.690
|
NM_016719
|
Grb14
|
growth factor receptor bound protein 14
|
|
chr4_+_140977621
|
29.651
|
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular)
|
|
chrX_-_92673432
|
29.173
|
NM_080858
|
Asb12
|
ankyrin repeat and SOCS box-containing 12
|
|
chr3_+_116265890
|
29.105
|
NM_027321
NM_175413
|
Lrrc39
|
leucine rich repeat containing 39
|
|
chr4_-_9570815
|
28.657
|
NM_001177852
NM_001177853
NM_001177854
NM_133723
|
Asph
|
aspartate-beta-hydroxylase
|
|
chr1_+_137695997
|
28.564
|
NM_021467
NM_001112702
|
Tnni1
|
troponin I, skeletal, slow 1
|
|
chr3_+_137004089
|
28.150
|
|
Emcn
|
endomucin
|
|
chr16_-_85455370
|
28.145
|
|
|
|
|
chr2_+_120289460
|
27.913
|
|
Capn3
|
calpain 3
|
|
chr19_-_40476799
|
27.611
|
NM_001034964
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr3_-_142058569
|
27.526
|
|
Pdlim5
|
PDZ and LIM domain 5
|
|
chr5_-_130354822
|
27.183
|
NM_011079
|
Phkg1
|
phosphorylase kinase gamma 1
|
|
chr16_+_45093784
|
27.082
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr2_+_120289326
|
27.049
|
NM_001177799
NM_007601
|
Capn3
|
calpain 3
|
|
chr8_-_25549415
|
26.752
|
NM_026931
|
1810011O10Rik
|
RIKEN cDNA 1810011O10 gene
|
|
chr3_+_32716447
|
26.686
|
NM_001013024
|
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr8_-_25549374
|
26.563
|
|
1810011O10Rik
|
RIKEN cDNA 1810011O10 gene
|
|
chr4_+_135803834
|
26.352
|
NM_011542
|
Tcea3
|
transcription elongation factor A (SII), 3
|
|
chr3_+_137004041
|
26.172
|
NM_001163522
NM_016885
|
Emcn
|
endomucin
|
|
chr1_-_45559700
|
25.291
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr6_+_112223751
|
25.257
|
NM_144799
|
Lmcd1
|
LIM and cysteine-rich domains 1
|
|
chr7_-_123174560
|
25.079
|
NM_001025560
|
Sox6
|
SRY-box containing gene 6
|
|
chr2_-_76820599
|
24.860
|
NM_011652
NM_028004
|
Ttn
|
titin
|
|
chr9_+_121686724
|
24.814
|
NM_028202
|
Kbtbd5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr7_+_52661329
|
24.120
|
NM_010596
|
Kcna7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr5_-_17394564
|
23.664
|
|
Cd36
|
CD36 antigen
|
|
chr3_-_10208575
|
23.631
|
NM_024406
|
Fabp4
|
fatty acid binding protein 4, adipocyte
|
|
chr6_-_97202685
|
23.565
|
NM_001081157
|
Lmod3
|
leiomodin 3 (fetal)
|
|
chr8_-_47296201
|
23.403
|
NM_007450
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4
|
|
chr11_-_120509650
|
23.184
|
NM_026751
|
Myadml2
|
myeloid-associated differentiation marker-like 2
|
|
chr15_+_31498611
|
23.180
|
NM_181588
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas)
|
|
chr1_-_45560001
|
22.906
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr7_-_31839825
|
22.172
|
NM_052991
NM_194321
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1
|
|
chr8_+_24168730
|
22.061
|
NM_031158
|
Ank1
|
ankyrin 1, erythroid
|
|
chr10_+_126938744
|
22.046
|
NM_177707
|
Stac3
|
SH3 and cysteine rich domain 3
|
|
chr8_+_122190264
|
21.762
|
NM_023395
|
Wfdc1
|
WAP four-disulfide core domain 1
|
|
chr1_+_137732910
|
21.534
|
|
Tnnt2
|
troponin T2, cardiac
|
|
chr1_-_72920776
|
21.368
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr8_+_55635768
|
21.176
|
|
Asb5
|
ankyrin repeat and SOCs box-containing 5
|
|
chr13_-_115078961
|
21.092
|
|
Ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4
|
|
chr2_+_31951149
|
20.438
|
NM_145521
|
Ppapdc3
|
phosphatidic acid phosphatase type 2 domain containing 3
|
|
chr5_-_69933835
|
20.358
|
NM_023784
|
Yipf7
|
Yip1 domain family, member 7
|
|
chr1_+_75356906
|
20.351
|
|
Des
|
desmin
|
|
chr10_-_62666659
|
19.901
|
NM_182992
|
Mypn
|
myopalladin
|
|
chr16_+_43640198
|
19.851
|
|
Zbtb20
|
zinc finger and BTB domain containing 20
|
|
chr11_+_72974053
|
19.423
|
|
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5
|
|
chr10_+_80657435
|
19.318
|
|
2310050B05Rik
|
RIKEN cDNA 2310050B05 gene
|
|
chr14_-_104213981
|
19.276
|
|
|
|
|
chr6_-_71212223
|
19.098
|
NM_001160127
NM_009762
|
Smyd1
|
SET and MYND domain containing 1
|
|
chr8_+_46744023
|
19.079
|
|
Sorbs2
|
sorbin and SH3 domain containing 2
|
|
chr15_-_41700964
|
19.038
|
NM_175456
|
Abra
|
actin-binding Rho activating protein
|
|
chr2_+_120302644
|
18.866
|
NM_001109761
|
Capn3
|
calpain 3
|
|
chr12_+_37835132
|
18.854
|
NM_008584
|
Meox2
|
mesenchyme homeobox 2
|
|
chr12_+_70342818
|
18.770
|
NM_178253
|
Klhdc1
|
kelch domain containing 1
|
|
chr10_-_80664743
|
18.753
|
NM_027120
|
Itgb1bp3
|
integrin beta 1 binding protein 3
|
|
chr7_-_104566013
|
18.704
|
NM_009381
|
Thrsp
|
thyroid hormone responsive SPOT14 homolog (Rattus)
|
|
chr1_-_137268058
|
18.680
|
|
Shisa4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr14_+_66587319
|
18.318
|
NM_013492
|
Clu
|
clusterin
|
|
chr1_+_75371871
|
18.201
|
NM_007463
|
Speg
|
SPEG complex locus
|
|
chr1_+_166726827
|
17.846
|
NM_019759
|
Dpt
|
dermatopontin
|
|
chr18_-_12914177
|
17.351
|
|
Osbpl1a
|
oxysterol binding protein-like 1A
|
|
chr13_-_62959656
|
17.341
|
NM_007994
|
Fbp2
|
fructose bisphosphatase 2
|
|
chr6_+_112409684
|
17.332
|
|
Cav3
|
caveolin 3
|
|
chr9_-_77195596
|
17.207
|
NM_027150
|
2310046A06Rik
2810047F03Rik
|
RIKEN cDNA 2310046A06 gene
RIKEN cDNA 2810047F03 gene
|
|
chr17_-_14831058
|
17.113
|
|
Thbs2
|
thrombospondin 2
|
|
chr3_+_137949734
|
16.746
|
|
Adh1
|
alcohol dehydrogenase 1 (class I)
|
|
chr3_-_10208480
|
16.708
|
|
Fabp4
Fabp9
|
fatty acid binding protein 4, adipocyte
fatty acid binding protein 9, testis
|
|
chr3_-_129458190
|
16.655
|
NM_010113
|
Egf
|
epidermal growth factor
|
|
chr14_+_48268985
|
16.446
|
|
Ktn1
|
kinectin 1
|
|
chr17_-_14831095
|
16.414
|
|
Thbs2
|
thrombospondin 2
|
|
chr13_-_62959751
|
16.055
|
|
Fbp2
|
fructose bisphosphatase 2
|
|
chr7_-_106494110
|
16.026
|
|
|
|
|
chr9_-_79825502
|
15.979
|
NM_001081243
|
Filip1
|
filamin A interacting protein 1
|
|
chr9_+_54442900
|
15.961
|
|
Idh3a
|
isocitrate dehydrogenase 3 (NAD+) alpha
|
|
chr4_+_48676385
|
15.952
|
NM_026509
|
Murc
|
muscle-related coiled-coil protein
|
|
chr11_-_101285982
|
15.948
|
|
1700113I22Rik
|
RIKEN cDNA 1700113I22 gene
|
|
chr11_-_69211724
|
15.688
|
NM_025915
|
Tmem88
|
transmembrane protein 88
|
|
chr5_-_17341512
|
15.234
|
|
Cd36
|
CD36 antigen
|
|
chr4_-_108311676
|
15.164
|
|
|
|
|
chr11_+_67090625
|
15.082
|
NM_177369
|
Myh8
|
myosin, heavy polypeptide 8, skeletal muscle, perinatal
|
|
chr5_+_25265761
|
15.060
|
NM_001004365
|
Actr3b
|
ARP3 actin-related protein 3 homolog B (yeast)
|
|
chr15_+_73578771
|
14.829
|
|
Ptp4a3
|
protein tyrosine phosphatase 4a3
|
|
chr1_-_72921438
|
14.816
|
NM_010518
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr17_+_75835208
|
14.809
|
NM_001166493
|
Rasgrp3
|
RAS, guanyl releasing protein 3
|
|
chr6_+_116600624
|
14.618
|
NM_001166580
NM_145980
|
8430408G22Rik
|
RIKEN cDNA 8430408G22 gene
|
|
chr9_-_66880619
|
14.284
|
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr16_+_56477956
|
14.134
|
NM_001014399
NM_001014422
NM_001014423
NM_001014424
NM_178790
|
Abi3bp
|
ABI gene family, member 3 (NESH) binding protein
|
|
chr4_+_104829951
|
13.914
|
NM_080555
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr2_-_62411970
|
13.912
|
NM_007986
|
Fap
|
fibroblast activation protein
|
|
chr10_-_24919916
|
13.757
|
|
Akap7
|
A kinase (PRKA) anchor protein 7
|
|
chr5_-_124053691
|
13.678
|
|
Clip1
|
CAP-GLY domain containing linker protein 1
|
|
chr1_+_137732961
|
13.662
|
NM_001130174
NM_001130175
NM_001130177
NM_001130178
NM_001130179
NM_001130180
NM_001130181
NM_011619
NM_001136083
|
Tnnt2
|
troponin T2, cardiac
|
|
chr7_+_120909625
|
13.572
|
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein
|
|
chr2_-_125142715
|
13.541
|
|
Fbn1
|
fibrillin 1
|
|
chr16_+_45093816
|
13.464
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr6_+_4471231
|
13.431
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr4_+_57907032
|
13.431
|
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr11_+_70470701
|
13.426
|
NM_001136062
|
Eno3
|
enolase 3, beta muscle
|
|
chr19_-_37250472
|
13.408
|
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr3_-_122939013
|
13.375
|
NM_080451
|
Synpo2
|
synaptopodin 2
|
|
chr11_+_94189524
|
13.341
|
|
Ankrd40
|
ankyrin repeat domain 40
|
|
chr2_+_31951030
|
13.329
|
|
Ppapdc3
|
phosphatidic acid phosphatase type 2 domain containing 3
|